How To Import Data Into Rstudio
Importing data and managing files
Learning objectives:
- Create projects in Rstudio
- Proper steps to import data
- Intro to data transformation using dplyr
Projection-oriented workflow
Information technology is best to create a split up folder that contains all related files. Yous can exercise the same for research projects. In Rstudio, this is called a Project. For example you can create a project for each of the chapters in this book. Projects are cocky-contained and tin be moved to some other reckoner or folder and the code will run.
If the first line of your R script is
setwd("C:\Users\jenny\path\that\just\I\have")
I will come into your office and SET YOUR Reckoner ON Burn
—@JennyBryan on Twitter

Animated GIF for creating projection and import information.
Create a projection in a new folder
Start by setting up a project in a new folder by going to File->New projection->New Directory->Empty Project. And so choose where the directory will be created on your hard drive. I created a directory chosen "Chapter4" under "D:\RBook." Rstudio creates a Project file named like "Chapter4.Rproj," which contains information such as scripts files and working folders. Projects files can exist saved and later on opened from File->Open up. In Windows you tin also just double-click on the project file. Y'all become everything ready to keep a item consignment or research project.
This folder is your working binder, a default folder for reading files, writing outputs etc.
More avant-garde users should consider create a projection using version command, so that all versions of your code is backup and easily shared past GitHub. Run across Happy Git and GitHub for the useR by Jenny Bryan.
Copy data files to the new directory
This is done outside of Rstudio on Windows or Mac interface. Download the heartatk4R.txt file from here. Information technology is a tab-delimited text file, meaning the dissimilar columns are separated by tab. We also demand to become some context virtually the data and how it is collected. At the minimum level, nosotros need to know the meaning of each columns.
File unzipping, conversion, and context. If data is compressed, unzip using seven-goose egg, WinRAR, Winzip, gzip. Any of these volition do. Is it a text file (CSV, txt, …) or Binary file (XLS, XLSX, …)? Convert binary to text file using corresponding application. Comma separated values (CSV) files, use comma to separate the columns. Another mutual type is tab-delimited text files, which uses the tab or \(\t\) as it is invisible grapheme.
Checking the file with text editor and Excel. Before reading files into R, we ofttimes demand to open the files to take a look. Notepad or WordPad that come with Windows is very limited (and then apprentice)! Do not even think about using Microsoft Discussion! I strongly recommend that yous install a powerful text editor such as NotePad++ (https://notepad-plus-plus.org/), or TextPad (https://www.textpad.com/). If you are a Mac user, attempt TextMate, TextWrangler etc. I utilise NotePad++ almost every day to look into data, and too write R programs, as it tin highlight R commands based on R syntax. I even use a tool called NppToR (https://sourceforge.net/projects/npptor/) to ship R commands from NotePad++ directly to R, and I beloved it!
Regardless of their extensions in file names, all apparently text files tin be opened by these text editors. Plain text files only contain text without any formatting, links and images. The file names tin can be "poems.txt," "poems.tex," "students.csv," or but "data" without extension. I often salvage my R scripts as text file with names like "code_1-22-2017.R." You can import text files, regardless of file names, to Microsoft Excel, which can properly parse your file into columns if the correct delimiter is specified.
Comma separated values (CSV) files utilize comma to split the columns. CSV files tin also be conveniently opened past Excel. And Rstudio likes it too. Then let's effort to utilise CSV files. Another common type is tab-delimited text files, which uses the tab or \(\t\) as it is invisible character. Other types of files such as Excel .xls or .xlsx files often needed to be saved every bit CSV files.
Import data files
In Rstudio, click File->Import Dataset->From text(readr)…, observe the file on your hard drive. You lot should alter the Delimiter to "tab," and the preview shows that the data is correctly parsed into multiple columns. Yous tin can likewise change the name of your data object by changing the default "heartatk4R" to "df" on the lower left of the import interface.
We need to bank check each of the columns and encounter the data types are correctly read in. The get-go column is just patient id number runs from 1 to 12844. Information technology will not exist useful in our analysis. The numbers in DIAGNOSIS, DRG, and DIED are integers but they really lawmaking for certain categories. They are not measurements. It does not make sense, for example, to add them or boilerplate them. Most of the times, there is no particular order. The same is true for SEX. So in this dialog interface, DIAGNOSIS, DRG, and DIED should be changed from "double" to "character". As shown in Effigy 4.2, y'all tin click on the automatically guessed information type under each of the column names as shown in Effigy 4.two. Past selecting "character" from the drop downwardly, y'all can successfully format this column every bit a character.
On the other mitt, LOS (length of stay in days) and Age should exist numbers. Only because 10 is presented as "0010," these columns are automatically recognized equally characters. Nosotros take to force R to read these columns every bit integers by clicking on the column championship and select integer. We have to change LOS and Historic period from "character" to "integer".
The crawly nerds at Rstudio really helped yous generating these 3 lines of code:
library(readr) df <- read_delim("datasets/heartatk4R.txt", " \t ", escape_double = Fake, col_types = cols(Historic period = col_integer(), DIAGNOSIS = col_character(), DIED = col_character(), DRG = col_character(), LOS = col_integer()), trim_ws = TRUE) View(df) Before yous click on the Import push, I highly recommend that you select all the codes and re-create it to the script file. Later on clicking Import, you tin can paste the code into a script window. If you exercise non have a script window open, y'all tin create one by clicking the File + icon on the peak left. Copy and paste these code to your script file. You lot will demand it when yous want to re-run the analysis without going through the above steps. You can run into the information appears as a spreadsheet, which can be sorted by clicking on the cavalcade names. This spreadsheet can exist closed. To reopen, click on df object, which is a data frame named after the input file. You information is now available as df.
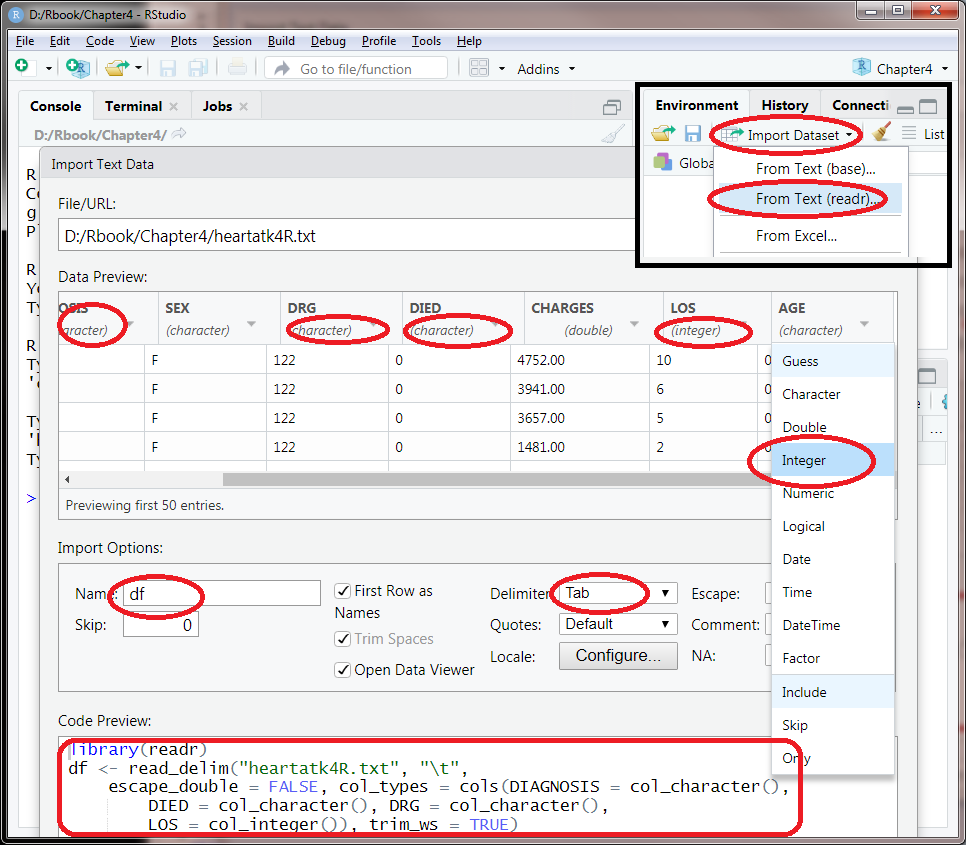
Figure four.2: Changing data types while importing data into Rstudio.
Check and convert data types
Most of the times, R can gauge the type of information in each column. But we always demand to double check using the str control. If not satisfied, we can enforce data blazon conversion in R using as.numeric, as.factor, or as.character functions.
str(df) # structure of data object, data types for each column ## spec_tbl_df [12,844 10 8] (S3: spec_tbl_df/tbl_df/tbl/data.frame) ## $ Patient : num [1:12844] 1 ii 3 4 5 6 vii 8 9 10 ... ## $ DIAGNOSIS: chr [1:12844] "41041" "41041" "41091" "41081" ... ## $ SEX : chr [i:12844] "F" "F" "F" "F" ... ## $ DRG : chr [i:12844] "122" "122" "122" "122" ... ## $ DIED : chr [ane:12844] "0" "0" "0" "0" ... ## $ CHARGES : num [1:12844] 4752 3941 3657 1481 1681 ... ## $ LOS : int [ane:12844] ten half dozen 5 2 1 9 xv fifteen 2 ane ... ## $ Age : int [1:12844] 79 34 76 lxxx 55 84 84 70 76 65 ... ## - attr(*, "spec")= ## .. cols( ## .. Patient = col_double(), ## .. DIAGNOSIS = col_character(), ## .. SEX = col_character(), ## .. DRG = col_character(), ## .. DIED = col_character(), ## .. CHARGES = col_double(), ## .. LOS = col_integer(), ## .. AGE = col_integer() ## .. ) ## - attr(*, "problems")=<externalptr> Nosotros can reformat chiselled values factors. Nosotros are going to utilise df$Sexual activity to refer to the Sex column of the data frame df:
df$DIAGNOSIS <- as.factor(df$DIAGNOSIS) # convert this column to factor df$Sex activity <- as.factor(df$SEX) df$DRG <- as.cistron(df$DRG) df$DIED <- as.factor(df$DIED) Factors are very similar to character vectors. Only factors have defined levels and their values are stored as integers.
## [i] ix ## [1] "41001" "41011" "41021" "41031" "41041" "41051" "41071" "41081" "41091" Annotation "41001" is the reference level for this factor and it is coded as 1. The reference level for factors are of import when we interpret results from regression, as effects are represented relative to the reference level. The reference level are defined based on the social club they appear in the dataset. Sometimes we demand to change the reference level.
df$DIAGNOSIS <- relevel( df$DIAGNOSIS, "41091") levels(df$DIAGNOSIS) ## [one] "41091" "41001" "41011" "41021" "41031" "41041" "41051" "41071" "41081" str(df) # double check structure of data ## spec_tbl_df [12,844 x 8] (S3: spec_tbl_df/tbl_df/tbl/data.frame) ## $ Patient : num [1:12844] i 2 iii four 5 half dozen 7 8 9 10 ... ## $ DIAGNOSIS: Factor w/ 9 levels "41091","41001",..: 6 6 i 9 1 ane 1 i 6 half-dozen ... ## $ SEX : Factor west/ ii levels "F","M": ane i ane one 2 2 1 one 2 1 ... ## $ DRG : Cistron westward/ three levels "121","122","123": 2 2 two ii 2 1 1 1 1 3 ... ## $ DIED : Factor w/ 2 levels "0","1": 1 i 1 1 ane ane 1 1 1 2 ... ## $ CHARGES : num [1:12844] 4752 3941 3657 1481 1681 ... ## $ LOS : int [one:12844] 10 6 five 2 ane 9 15 15 2 i ... ## $ AGE : int [ane:12844] 79 34 76 80 55 84 84 seventy 76 65 ... ## - attr(*, "spec")= ## .. cols( ## .. Patient = col_double(), ## .. DIAGNOSIS = col_character(), ## .. Sexual practice = col_character(), ## .. DRG = col_character(), ## .. DIED = col_character(), ## .. CHARGES = col_double(), ## .. LOS = col_integer(), ## .. Historic period = col_integer() ## .. ) ## - attr(*, "problems")=<externalptr> summary(df) # a summary oftentimes gives us a lot of useful information ## Patient DIAGNOSIS Sex activity DRG DIED CHARGES ## Min. : 1 41091 :5213 F:5065 121:5387 0:11434 Min. : 3 ## 1st Qu.: 3212 41041 :2665 M:7779 122:6047 ane: 1410 1st Qu.: 5422 ## Median : 6422 41011 :1824 123:1410 Median : 8445 ## Mean : 6422 41071 :1703 Hateful : 9879 ## 3rd Qu.: 9633 41001 : 467 3rd Qu.:12569 ## Max. :12844 41081 : 287 Max. :47910 ## (Other): 685 NA's :699 ## LOS Age ## Min. : 0.000 Min. : 20.00 ## 1st Qu.: iv.000 1st Qu.: 57.00 ## Median : 7.000 Median : 67.00 ## Mean : seven.569 Mean : 66.29 ## third Qu.:ten.000 3rd Qu.: 77.00 ## Max. :38.000 Max. :103.00 ## The summary( ) function is very useful to get bones information about data frames.
Note that for numeric columns we are shown mean, median, etc, while for factors the frequencies are shown. This reassured u.s.a. that the data types are correctly recognized. It besides shows missing values for CHARGES. Some people got free treatment for center attack? Perchance not. Missing does not mean zippo. Perhaps the data was not entered for some patients.
Except enforcing data blazon conversion by as.factor, as.numeric and then on, We can also reformat the columns before clicking Import as we described in iv.two.4.
Shut a project when yous are done
Once you lot are done with a project, you can close information technology from File \(\rightarrow\)Shut Project. If you do not close a project, Rstuio assumes that you will be continue working on the same project; even after your close Rstudio, the same project and files will be open adjacent time when you lot offset Rstudio. This is convenient, virtually of times. Only I've seen some students' Rstudio environment cluttered with all the things they have been doing for an entire semester. This is not just confusing, only could also lead to errors. For example in this form, yous can start a new project for each chapter.
To open a project, use File \(\rightarrow\)Open Project and and then navigate to the project. Alternatively you can double-click on the Chapter4.Rproj file from Windows or Mac. When a project file is loaded, the entire computing envirionment is set for you lot. The working directory is gear up properly. Some of the script files are open up. If the script file is not open, you tin open up it by clicking on it from the Files tab in the lower right window.
Reading files directly using read.tabular array
As you become more feel with R programming, there are many other options to import data, except using the Import Dataset in Rstudio.
In summary, we have the following code to read in the heart assail dataset. We accept to brand sure the file is in the current working directory. To fix working directory from Rstudio primary carte, get to Session -> Gear up Working Directory.
rm(list = ls()) # Erase all objects in retentivity getwd() # show working directory df <- read.table("datasets/heartatk4R.txt", sep= " \t ", header = TRUE) head(df) # show the first few rows # change several columns to factors df$DRG <- equally.factor(df$DRG) df$DIED <- every bit.factor(df$DIED) df$DIAGNOSIS <- as.gene(df$DIAGNOSIS) df$Sexual practice <- equally.factor(df$SEX) str(df) # testify the data types of columns summary(df) # show summary of dataset Alternatively, you can skip all of the to a higher place and practice this.
URL <- "https://raw.githubusercontent.com/gexijin/learnR/master/datasets/heartatk4R.txt" df <- read.table(URL, header = Truthful, sep = " \t ", colClasses = c("grapheme", "factor", "factor", "factor", "gene", "numeric", "numeric", "numeric")) We are reading information direct from the internet with the URL. And we are specifying the data type for each column.
General procedure to read data into R:
- If information is compressed, unzip using 7-zip, WinRAR, Winzip, gzip. Any of these will do.
- Is information technology a text file (CSV, txt, …) or Binary file (XLS, XLSX, …)? Convert binary to text file using corresponding application. Comma separated values (CSV) files use comma to separate the columns. Some other mutual type is tab-delimited text files, which uses the tab or \(\t\) as it is invisible character.
- Open with a text editor (TexPad, NotePad++) to have a look.
- Rows and columns? Row and cavalcade names? row.names = 1, header = T
- Delimiters betwixt columns?(space, comma, tab…) sep = "\(\t\)"
- Missing values? NA, na, Null, bare, NaN, 0 missingstring =
- Open equally text file in Excel, cull advisable delimiter while importing, or use the Text to Column under Data in Excel. Beware of the abrasive automatic conversion in Excel "OCT4"->"four-OCT." Edit column names by removing spaces, or shorten them for piece of cake of reference in R. Save every bit CSV for reading in R.
- read.table( ), or read.csv( ). For example,
x <- read.table("somefile.txt", sep = "\t", header = TRUE, missingstring = "NA") - Double check the data with str(ten), make sure each column is recognized correctly every bit "character", "gene" and "numeric". Pay attention to columns comprise numbers but are actually IDs (i.e. pupil IDs), these should exist treated as character. For example,
10$ids <- as.character(x$ids), here x is the data frame and ids is the column name. Also pay attention to columns contain numbers but actually codes for some discrete categories (i, ii, three, representing treatment 1, treatment ii and handling 3). These demand to be reformatted as factors. This could exist done with something similarx$treatment <- as.factor(x$handling).
Refresher using cheat sheets that summarize many R functions is available here: https://www.rstudio.com/resources/cheatsheets/. It is of import to know the unlike types of R objects: scalars, vectors, data frames, matrix, and lists.
Practise 4.i
If y'all accept not created a projection for chapter four, it is fourth dimension to create i. Download the tab-delimited text file pulse.txt from this page (http://statland.org/R/R/pulse.txt). Import pulse.txt into R using two methods: R menu (Show the process by attaching some necessary screenshots.) and R script.
a. Rename the file as chapter4Pulse.
b. Modify the class of ActivityL from double to integer.
c. After importing pulse.txt into R, convert the class of Sex activity from charater to cistron using R lawmaking. Don't forget using class() function to check your respond.
Exercise 4.2
Type in Tabular array 4.1 in Excel and salvage as a CSV file and a tab-delimited tex file. Create a new Rstudio project equally outlined above. Copy the files to the new folder. Import the CSV file to Rstudio. Create a script file which includes the rm(list = ls()) and getwd() command, the generated R code when importing the CSV file, (similar to those shown in Figure 4.ii), and the code that convert data types (Historic period, BloodPressure and Weight should be numeric, LastName should be character and HeartAttack should exist factor). Name the data prepare as patients. Submit the R script your created, data structure of the data set patient, and use caput(patients) to prove the data.
| LastName | Historic period | Sex | BloodPressure | Weight | HeartAttack |
|---|---|---|---|---|---|
| Smith | xix | M | 100 | 130.ii | 1 |
| Bird | 55 | F | 86 | 300 | 0 |
| Wilson | 23 | M | 200 | 212.vii | 0 |
Enter data manually
There are many unlike ways to go information into R. Yous can enter data manually (see below), or semi-manually (see below). You can read data into R from a local file or a file on the net. You can also use R to recollect data from databases, local or remote. The most important thing is to read data set into R correctly. A dataset not read in correctly will never be analyzed or visualized correctly.

x <- c(2.1, 3.1, 3.2, five.iv) sum(ten) ## [ane] 13.8 A <- matrix( c(2, 4, 3, one, v, 7), # the data elements nrow = 2, # number of rows ncol = three) # number of columns A # show the matrix ## [,1] [,two] [,3] ## [1,] 2 3 v ## [2,] four i vii x <- scan() # Enter values from keyboard, separated by Return central. End by empty line. 2.i ## [1] ii.1 ## [1] 3.1 ## [1] 4.1 You can fifty-fifty use the scan() function, and so copy and paste a cavalcade of numbers from Excel.
Information manipulation in a information frame
For our heartattach dataset df, nosotros tin can sort the information by age. Again, type these commands in the script window, instead of directly into the Panel window. And salve the scripts in one case a while.
df2 <- df[order(df$Historic period), ] # sort by ascending order by Age Global Surround window contains the names and sizes of all the variables or objects in the calculator memory. R programming is all about creating and modifying these objects in the memory with clear, footstep-by-step instructions. We also can sort the data by clicking on the column names in spreadsheet from Global Environs.
Just like in Excel, you can add a new column with computed results:
df2$pdc <- df2$CHARGES / df2$LOS Here we created a new column pdc to represent per day cost. We can also create a column to correspond age groups using the floor function just returns the integer part.
df2$ag <- floor(df2$Historic period/ 10) * 10 You can now do things like this:
boxplot(df2$CHARGES ~ df2$ag) 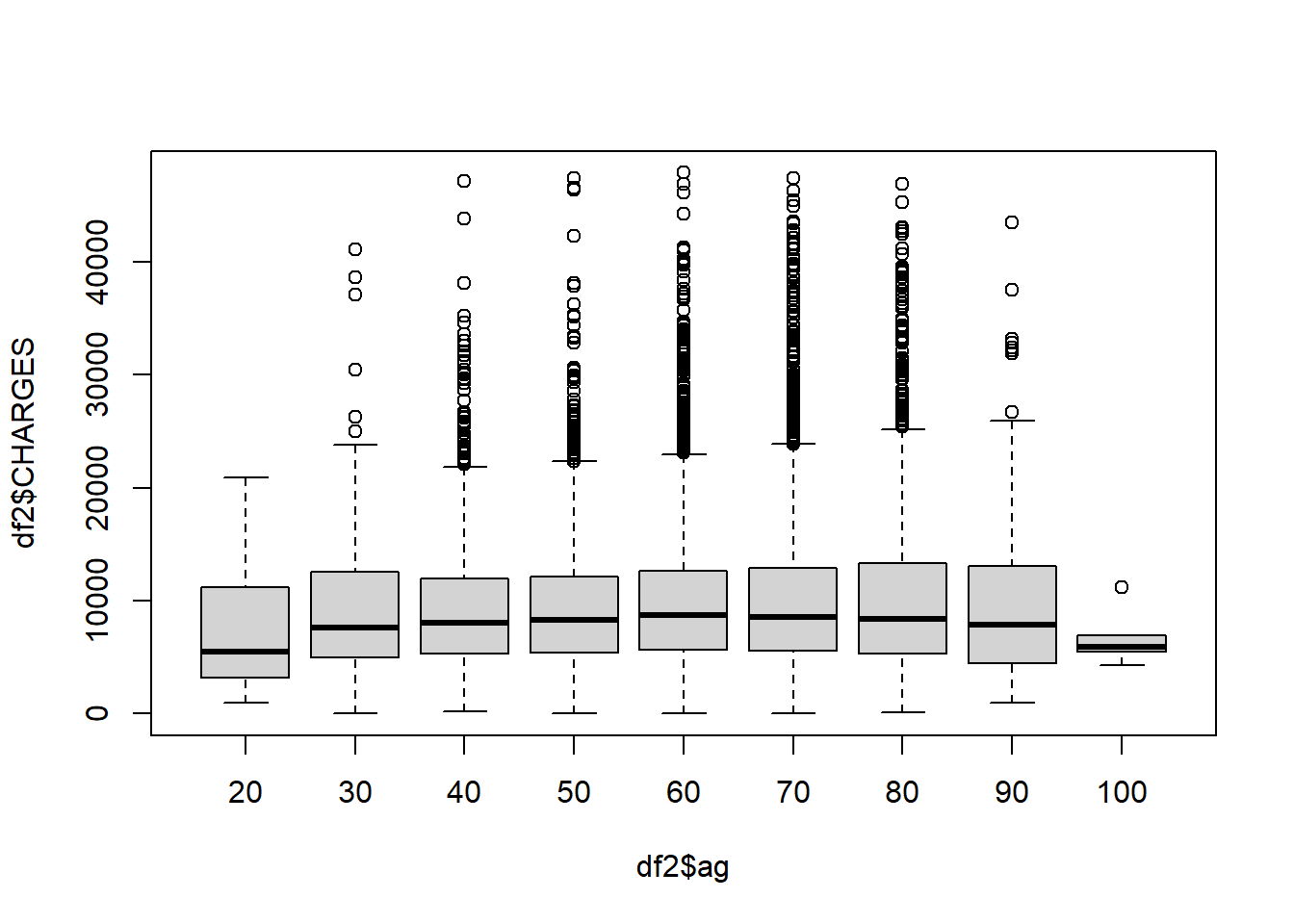
Each box represents an age grouping. Older patients tends to stay longer in the hospital after beingness admitted for eye attack.
You can extract a subset of cases:
df3 <- subset(df2, Sex activity == "F") # But females. "==" is for comparison and "=" is for assign value. df4 <- subset(df3, Historic period > 80) # only people older than 80 summary(df4) ## Patient DIAGNOSIS SEX DRG DIED CHARGES ## Min. : 7 41091 :564 F:1263 121:679 0:929 Min. : 92 ## 1st Qu.: 3416 41071 :224 G: 0 122:250 i:334 1st Qu.: 5179 ## Median : 6816 41041 :181 123:334 Median : 8580 ## Hateful : 6624 41011 :166 Mean :10143 ## 3rd Qu.: 9814 41001 : 41 third Qu.:13347 ## Max. :12841 41081 : 38 Max. :46915 ## (Other): 49 NA's :74 ## LOS AGE pdc ag ## Min. : ane.000 Min. : 81.00 Min. : xviii.four Min. : lxxx.00 ## 1st Qu.: 5.000 1st Qu.: 83.00 1st Qu.: 796.7 1st Qu.: eighty.00 ## Median : 8.000 Median : 85.00 Median : 1113.7 Median : 80.00 ## Hateful : 9.457 Mean : 85.92 Hateful : 1331.4 Mean : 81.96 ## tertiary Qu.:13.000 third Qu.: 89.00 3rd Qu.: 1523.viii 3rd Qu.: 80.00 ## Max. :38.000 Max. :102.00 Max. :11246.i Max. :100.00 ## NA's :74 Attempt not to attach the data when you are manipulation data like this.
Data transformation using the dplyr
Following the same style of ggplot, the dplyr package, a part of the Tidyverse, makes data transformation more intuitive.
Get-go we demand to install the dplyr package.
install.packages("dplyr") With dplyr, data transformation becomes fun.
library(dplyr) df2 <- df %>% # piping operator; data is transport to the next footstep suit(Historic period) # sort in ascending gild; desc(AGE) for descending social club head(df2) ## Patient DIAGNOSIS SEX DRG DIED CHARGES LOS AGE ## 1 5411 41041 M 122 0 6214.00 iv 20 ## 2 10853 41091 F 122 0 6726.27 4 21 ## iii 4126 41041 M 122 0 10781.00 8 23 ## iv 10738 41011 1000 121 0 NA eight 23 ## v 4247 41091 F 122 0 10672.00 vi 24 ## vi 5199 41041 M 121 0 7596.00 viii 24 df2 <- df2 %>% mutate( pdc = CHARGES / LOS) # new column by dividing two columns head(df2) ## Patient DIAGNOSIS SEX DRG DIED CHARGES LOS Historic period pdc ## i 5411 41041 M 122 0 6214.00 4 twenty 1553.500 ## ii 10853 41091 F 122 0 6726.27 iv 21 1681.568 ## 3 4126 41041 M 122 0 10781.00 8 23 1347.625 ## 4 10738 41011 M 121 0 NA viii 23 NA ## v 4247 41091 F 122 0 10672.00 6 24 1778.667 ## 6 5199 41041 K 121 0 7596.00 viii 24 949.500 df2 <- df2 %>% mutate( ag = floor(Age/ ten) * x) # new column, age grouping caput(df2) ## Patient DIAGNOSIS Sexual practice DRG DIED CHARGES LOS AGE pdc ag ## 1 5411 41041 One thousand 122 0 6214.00 4 20 1553.500 xx ## two 10853 41091 F 122 0 6726.27 4 21 1681.568 20 ## 3 4126 41041 M 122 0 10781.00 8 23 1347.625 20 ## 4 10738 41011 Thou 121 0 NA viii 23 NA twenty ## 5 4247 41091 F 122 0 10672.00 vi 24 1778.667 20 ## 6 5199 41041 M 121 0 7596.00 8 24 949.500 20 df3 <- df %>% filter(Sex == "F", Historic period > eighty) # filtering head(df3) ## Patient DIAGNOSIS Sex DRG DIED CHARGES LOS Age ## 1 seven 41091 F 121 0 10958.52 xv 84 ## 2 13 41091 F 122 0 NA 9 83 ## 3 27 41011 F 123 1 3214.90 four 83 ## 4 36 41041 F 121 0 2584.x 9 81 ## 5 51 41011 F 121 0 7589.00 11 81 ## 6 67 41011 F 123 ane 16428.80 eight 81 The existent power and efficiency of dplyr comes when yous connect the pipes to exercise information transformation in multiple steps.
df2 <- df %>% arrange(AGE) %>% mutate( pdc = CHARGES / LOS) %>% mutate( ag = floor(AGE/ 10) * 10) %>% filter(Sex activity == "F", Age > fourscore) head(df2) ## Patient DIAGNOSIS SEX DRG DIED CHARGES LOS Historic period pdc ag ## one 36 41041 F 121 0 2584.10 ix 81 287.1222 80 ## 2 51 41011 F 121 0 7589.00 11 81 689.9091 80 ## 3 67 41011 F 123 1 16428.80 8 81 2053.6000 lxxx ## 4 225 41041 F 123 i 4674.l vii 81 667.7857 80 ## v 265 41091 F 122 0 9401.05 9 81 1044.5611 80 ## 6 284 41041 F 121 0 7240.30 seven 81 1034.3286 lxxx arrange, mutate, filter are called activity verbs. For more activeness verbs, see dplyr cheat sheet from the Rstudio main menu: Aid \(\rightarrow\) Cheatsheets \(\rightarrow\) R Markdown Crook Sheet. Information technology is also available on line dplyr cheat Sheet.
How To Import Data Into Rstudio,
Source: https://gexijin.github.io/learnR/importing-data-and-managing-files.html
Posted by: donohueyoulle64.blogspot.com


0 Response to "How To Import Data Into Rstudio"
Post a Comment